Overview
Abstract
The BioGear® Circuit Solver was created to extract the lumped parameter physics calculations from within the individual systems and centralize the generic calculations. Closed loop circuits can be defined easily within the Common Data Model (CDM). They are fully dynamic, and can be manipulated and modified at each time step. The solver can be used to analyze electrical, fluid, and thermal circuits using native units. The solver outputs were validated using the outputs from an existing known third party circuit solver. All results matched the validation data, confirming this is a sound approach for the BioGears Engine.
Introduction
BioGears systems use lumped parameter circuits to mimic the physiology of the human body. These circuits use fluid or thermal elements that are analogous to electrical circuit elements. The circuits have several types of feedback mechanisms that can be set and changed at every time step. Figure 1 presents a generic example of very low fidelity lumped parameter physiology circuits. Circuits can be thought of as pipe networks for fluid analysis.
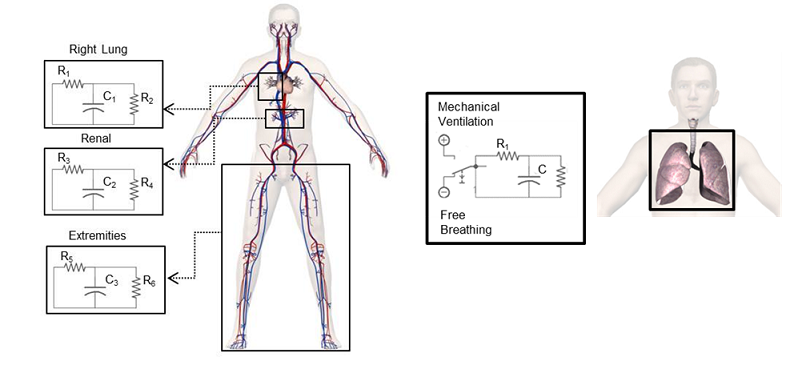
Note: For simplicity, this document uses electrical components and terminology when discussing the solver functionality. See Table 1 for analogies and details about the mapping of electrical components.
The BioGears CDM includes many of the same generic definitions traditionally used to define and analyze circuits. Paths are ideal conductor branches that may contain elements (i.e., resistors, capacitors, inductors, diodes, etc.). Nodes are junctions at the intersection of paths. Figure 2 shows these base circuit element definitions. Paths are each assigned one source and one target node. We use the convention of positive current from source to target when performing calculations.
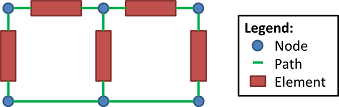
System Design
Background and Scope
Legacy Solution
The physiology engine from which BioGears was spawned (released by Advanced Simulation Corporation as Body Simulation for Anesthesia™) solved each lump parameter circuit using non-closed loop calculations contained within each system. The systems were extensively interconnected and the circuit calculations were unable to be decoupled. This made it extremely difficult to find sources of errors and inconsistencies in output results.
The circuit math was performed often using different approaches, with mixed results. It became apparent that neither energy nor mass was being truly conserved. This was true within the individual systems, as well as within the entire body. One-way fluid valves in the Cardiovascular System (similar to electrical diodes) would inadvertently allow backflow for a single time step when flows changed direction. Little validation was done to provide a sound fundamental base on which to build physiological systems.
Requirements
Given the overall goals of BioGears, we set out to create a generic circuit solver on which to expand. The solver is required to efficiently solve transient linear and non-linear circuits. Some other high-level requirements include:
- Generic - All systems should be able to use the same basic circuit physics engine. This allows rapid development, and makes engine outputs much easier to validate and verify.
- Computational Speed - BioGears is required to maintain a transient full-body solution faster than real time on typical personal computers.
- Modular - Using the same basis for design and construction will aid in keeping the system decoupled.
- Extensible - We must take future growth into consideration and allow users and developers the proper tools and building blocks on which to add new functionality.
- Dynamic - Feedback mechanisms are required for each system. It is beneficial to be able to dynamically change, add, or remove circuits, sub-circuits, elements, and connections at run time.
- Conservation - We must uphold sound scientific principles and conserve energy, mass, and momentum.
- Common Data Model - The entire solution must reside within and effectively use the Common Data Model.
- Circuits types - BioGears will include fluid and thermal circuits. It is beneficial to use the same solver for both types of systems.
- Elements - The solver must be able to handle all base circuit elements (i.e., resistors, capacitors, inductors, etc.), with room for expansion.
- Open source - As a whole, any third-party software must adhere to the same license requirements as BioGears.
Solutions Investigated
We considered several circuit solver options, including investigating existing software to determine if any would meet our needs. The best external code we found for integration into our C++ open source code was a variation on SPICE (Berkeley University’s Spice3f5), called Ngspice (http://ngspice.sourceforge.net/). Ngspice is an open source, general purpose circuit simulation program for nonlinear and linear analyses. It is developed with a C++ wrapper that maintains the original parametric netlist format (netlists can contain parameters and expressions) as inputs.
During implementation, we found Ngspice to be a good fit for our needs to solve closed-loop circuits, yet several negatives forced us to deviate from that path. We discovered a small number of bugs, some with possible workarounds. It was found to be very slow for our approach of solving a new (i.e., dynamic) circuit each time step. It was cumbersome to parse using the netlist input and output style. The most significant downside against implementing Ngspice into BioGears was the unavoidable build-up of memory that caused our software to crash.
Due to the aforementioned complications, we ultimately determined that creating our own generalized circuit analysis code was ideal and less labor-intensive than implementing existing software. This has the added benefit of allowing us to control every aspect of the analysis.
Data Flow
Our generic Circuit Solver intentionally mirrors the same data flow found in each of the systems. Each time step consists of a Preprocess call to set up the circuits for analysis, a Process call to do the analysis, and a Post Process call to advance the time in preparation for the next time step.
Preprocess
There is no literal Preprocess call within the circuit solver class (i.e., code). Each system individually and directly modifies its circuit(s). This allows our functionality to be entirely dynamic by performing a separate calculation every time step. See the Data Model Implementation Appendix below for details about BioGears circuit elements and the data they contain.
Process
For the circuit-solving software to effectively calculate the nonlinear circuit states without human intervention, a robust, generic, and repeatable methodology is required. The solver performs numerical integration through linearization (first order approximations) by assuming a direct current (DC) solution for the given time step. The Modified Nodal Analysis (MNA) approach is used to determine the state of every node and path within the circuit. The steps for solving a circuit in a given time step are (also see Figure 3):
Perform numerical integration by using linearization (first order approximations) through MNA, using the matrix algebra equation:
\[Ax = b\]
Equation 1.
Where A is the matrix of constants, x is the vector of unknowns/variables, and b is the right side vector of knowns.
- Use Kirkoff’s Current Laws (KCL) (sum of the currents is zero at each node) to populate the A matrix and b vector. Table 2 shows the equations used for determining flows, where flows (F) are equivalent to currents, and pressures (P) are equivalent to voltages.
- Leverage the Eigen templated library (released by Tuxfamily) LU decomposition linear solver (FullPivLU) to solve for unknown variables (i.e., voltages and currents). This decomposition provides the generic approach to solving systems of linear equations, and represents an LU decomposition of any matrix with complete pivoting. The matrix A is decomposed as:
\[A=P^{-1} LUQ^{-1} \]
Equation 2.
Where L is unit-lower-triangular, U is upper-triangular, and P and Q are permutation matrices. This is a rank-revealing LU decomposition. The eigenvalues (diagonal coefficients) of U are sorted in such a way that any zeros are at the end.
Calculate remaining unknown currents using the Trapezoidal Rule, where applicable. For nonlinear elements from time a to b, this is:
\[\int _{a}^{b}f(x)dx\approx (b-a)\left[\frac{f(a)+f(b)}{2} \right] \]
Equation 3.
- Calculate diode currents using assumed open or closed switch states (cannot be solved directly). Iterate Steps 1–3 until a satisfactory solution is attained.
Calculate the change in charge (Q) across capacitors based on the capacitance (C) and voltage (V) change, and increment the total charge. The charge is conserved on the source and target nodes by incrementing/decrementing this amount. Selecting which node gains charge and which loses is done by the direction of the current in the path.
\[Q(t)=C*V(t)\]
Equation 4.
- Invalidate the current on any path the user has specified to ignore. Note that this is to prevent unwanted transport to the reference node (i.e., ground).
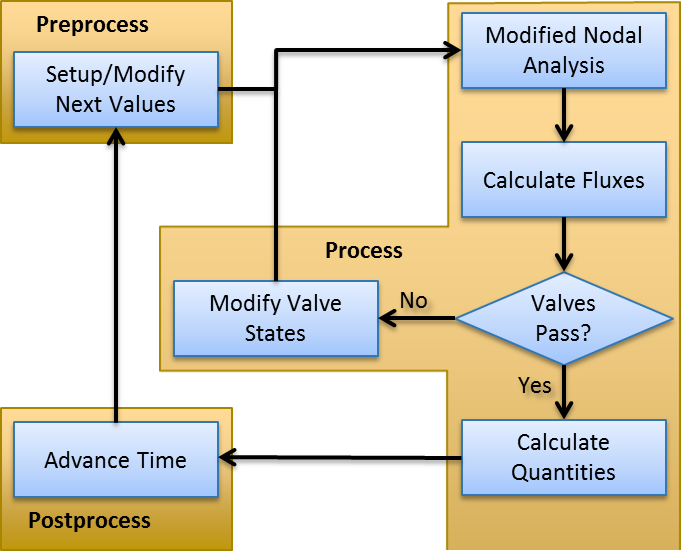
There are several nuances for the handling of certain elements:
- Paths without elements are calculated as if they are zero voltage sources, which allows for direct solving for the current.
- Switches are modeled as element-less paths when closed and essentially infinite resistances (10100 ohms) when open.
- Valves (i.e., Diodes) are modeled as closed switches when the path source node voltage is higher than the target node (on) and as open switches when the voltage is greater on the target node (off).
- Polarized elements are modeled as an open switch when preventing polarity reversal
Because diode and polarized element states cannot be determined before solving a circuit, we are forced to make a considered guess. Therefore, at each time step, we begin with an assumed on/off state for each of these elements in the circuit. We begin with the previous time step solution to minimize the number of iterations. The entire circuit is solved using this assumed state, and the currents and voltages associated with the paths are considered. When a diode/polarized element is found to have an incorrect current direction or voltage reversed relative to its assumed state, the state is flipped. This process is then repeated until the valid solution is found. Intelligently choosing which diode states to switch is important because there are 2^(Number of Diodes/Polarized Elements) possible combinations in each circuit.
Post Process
Post Process effectively advances time by moving the “next” time step parameter values to the “current” time step parameter values (see System Methodology).
Features and Capabilities
Approach and Extensibility
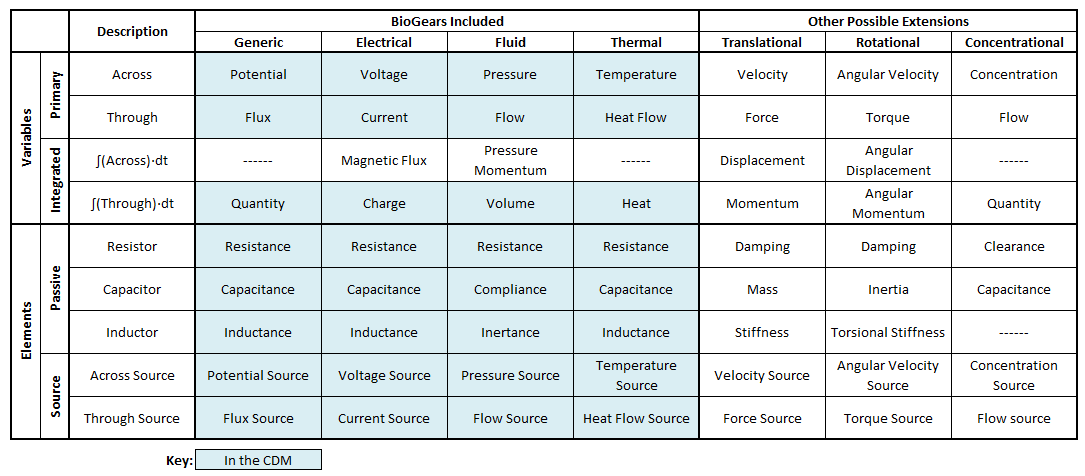
The BioGears systems use two different lumped parameter system types, each with analogies to electrical circuits. Most systems models (Cardiovascular, Respiratory, etc.) use fluid units (pressures, flows, and volumes), while others (Energy, Environment, etc.) use thermal units (temperature, heat transfer rate, and heat). Both model types use the same underlying physics to define relationships and solve for unknown values. We included the ability to calculate in native electrical units (voltage, current, and charge) to help with validation using known electrical circuit models and outputs. Table 1 lists system analogies and how they map to circuit elements.
Table 1. The various types of circuits that can be solved by the Circuit Solver. Some have units already defined and included in the CDM, while the others can be easily added. Possible inputs, outputs, elements, and parameters are all defined. Valves (diodes) and switches are not shown [283].
While we designed the generic Circuit Solver to analyze our fluid, thermal, and electrical models, we can easily extend it to include any of the model types in Table 1. Further details specific to the implementation of our model with the hydraulic analogy are shown in Table 2. A more intuitive pipe analogy is described through images. The BioGears defined fluid model elements are outlined in the first column. The flow equations are important for our analysis technique outlined earlier.
Table 2. In-depth description of the hydraulic analogy for electrical circuits that are used extensively inside BioGears. The Elements are defined by the CDM and used by the solver. The Flow equations are important for solving for the unknown parameters. [392]
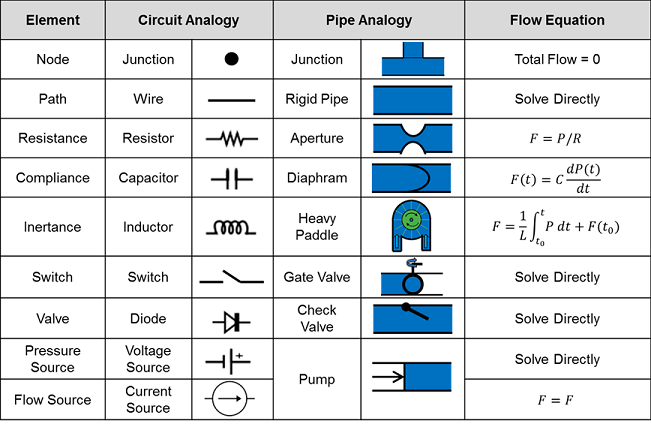
The Circuit Solver is equipped to use all of the element types given in the second column of Table 2. All three passive element types (resistor, capacitor, and inductor) have a polarized element modeling option. When active, polarized elements will short the circuit when the target node voltage becomes greater than that of the source node. This allows the user to model electrolytic capacitors and further ensures fluid will not be added to hydraulic systems if compliances switch polarity.
Data Model Implementation
Our mathematical approach to solving circuits is relatively straightforward, but our CDM implementation and integration with physiological models is novel. We implemented the Circuit Solver to use generic terms that are not specific to any one model type (see Table 1). Conversions to base units for each model are done in the background using the CDM unit conversion functionality. These base units are selected to prevent unnecessary conversion that would use critical computation resources, while still maintaining a direct mathematical relationship on both sides of the equation when performing the calculations.
The CDM allows us to implement the circuit math in a completely modular fashion. Every system uses the same physics, with the same CDM-defined elements. The burden on system developers is merely in setting up and manipulating the circuit correctly. A benefit to using a CDM is a significantly reduced development time.
The only significant requirement for circuit design is that it must be closed-loop. Beyond that, the solver can handle any combination of valid nodes, paths, and elements. Only one (or zero) element is allowed on each path and the convention of positive current going from source node to target node is used.
This sound foundation for defining and calculating circuit parameters, allows the engine to transport substances in a similarly generic fashion (see Drugs Methodology).
Assumptions
Circuits
There are several assumptions made for the circuit math in general:
- All circuits must be closed-loop
- All elements are idealized
- All circuits must have a reference node defined with a known voltage - unlike other circuit solvers, it does not necessarily have to have a potential value of zero
- Nonlinear sources need initial values. Therefore, paths with capacitors that do not have an initial voltage on the source or target nodes will assign it the reference node voltage value, and paths with inductors that do not have an initial current will be assigned a value of zero.
Fluids
There are other assumptions specific to the BioGears fluid systems:
- Flows are incompressible
- Fluids are inviscid
Limitations
There are frequency response limitations that are directly tied to the time step set externally to the solver itself. BioGears uses a time step of 1/50 s (~ 2 ms). Because the solver calculates nonlinear behavior through linearization using the trapezoid rule, extremely high frequency signal components will be lost. However, this far exceeds the Nyquist frequency for all current BioGears system models.
While it is not necessarily a limitation, users of the Circuit Solver must be careful to assign current and voltage source elements to set currents and voltages respectively. Attempting to directly set node voltages or path currents would result in an over-constrained solution. In these instances, the solver will overwrite those values.
Results and Conclusions
Validation
Beyond several numerical (hand) calculations used during implementation, we performed validation on the generalized circuit math by duplicating circuits in LTspice (version 4.21s, created by Linear Technology, Milpitas, CA). LTspice is based on the well-documented and validated SPICE simulator. We created circuits using all BioGears elements individually and in combination. We used several different types of dynamically-changing drivers to ensure proper transient functionality. The resulting voltage and current values were interpolated and validated to match for all 114 circuits. Table 3 shows a summary of the validation circuits investigated.
Table 3. The list of circuits created in BioGears and validated against LTspice. Every element is covered in combination with each other.
| Test Name | Purpose of test | Results Summary |
|---|---|---|
| BadDiodeDC | Intentionally failing to test Diode implementation | LTSpice: Will not run, BioGears: successfully fails - provides feedback on failure |
| BadDiodePULSE | Intentionally failing to test Diode implementation | LTSpice: Will not run, BioGears: successfully fails - provides feedback on failure |
| BadDiodeSIN | Intentionally failing to test Diode implementation | LTSpice: Will not run, BioGears: successfully fails - provides feedback on failure |
| BasicCircuit | Simple pressure source with resistances in series | Match |
| BasicDiodeDCCurrent | Diode behavior in response to a current source | Match |
| BasicDiodePULSECurrent | Diode behavior in response to a current source | Match |
| BasicDiodeSINCurrent | Diode behavior in response to a current source | Match |
| Comprehensive1DC | Combination of element types with a voltage source and a current source | Match |
| Comprehensive1PULSE | Combination of element types with a voltage source and a current source | Match |
| Comprehensive1SIN | Combination of element types with a voltage source and a current source | Match |
| CurrentCompDC | Combination of resistors and diode in response to a current source | Match |
| CurrentCompPulse | Combination of resistors and diode in response to a current source | Match |
| CurrentCompSIN | Combination of resistors and diode in response to a current source | Match |
| ParallelCapDC | Adding capacitors in parallel with a voltage source | Match |
| ParallelCapDCCurrent | Adding capacitors in parallel with a current source | Match |
| ParallelCapPulse | Adding capacitors in parallel with a voltage source | Match |
| ParallelCapPULSECurrent | Adding capacitors in parallel with a current source | Match |
| ParallelCapSIN | Adding capacitors in parallel with a voltage source | Match |
| ParallelCapSINCurrent | Adding capacitors in parallel with a current source | Match |
| ParallelCurrentSourceAdditionDC | Adding current sources in parallel | Match |
| ParallelCurrentSourceAdditionPULSE | Adding current sources in parallel | Match |
| ParallelCurrentSourceAdditionSIN | Adding current sources in parallel | Match |
| ParallelIndDC | Adding inductors in parallel with a voltage source | Match |
| ParallelIndDCCurrent | Adding inductors in parallel with a current source | Match |
| ParallelIndPULSE | Adding inductors in parallel with a voltage source | Match |
| ParallelIndPULSECurrent | Adding inductors in parallel with a current source | Match |
| ParallelIndSIN | Adding inductors in parallel with a voltage source | Match |
| ParallelIndSINCurrent | Adding inductors in parallel with a current source | Match |
| ParallelPressureSourceAdditionDC | Intentionally failing adding voltage sources in parallel | BioGears: successfully fails - provides feedback on failure |
| ParallelPressureSourceAdditionPULSE | Intentionally failing adding voltage sources in parallel | BioGears: successfully fails - provides feedback on failure |
| ParallelPressureSourceAdditionSIN | Intentionally failing adding voltage sources in parallel | BioGears: successfully fails - provides feedback on failure |
| ParallelRCDC | Parallel resistor and capacitor response to a voltage source | Match |
| ParallelRCPULSE | Parallel resistor and capacitor response to a voltage source | Match |
| ParallelRCSIN | Parallel resistor and capacitor response to a voltage source | Match |
| ParallelRDC | Parallel resistor response to a voltage source | Match |
| ParallelRDCCurrent | Parallel resistor response to a current source | Match |
| ParallelRLCDC | Parallel resistor, inductor, and capacitor response to a voltage source | Match |
| ParallelRLCDCCurrent | Parallel resistor, inductor and capacitor response to a current source | Match |
| ParallelRLCPULSE | Parallel resistor, inductor, and capacitor response to a voltage source | Match |
| ParallelRLCPULSECurrent | Parallel resistor, inductor and capacitor response to a current source | Match |
| ParallelRLCSIN | Parallel resistor, inductor, and capacitor response to a voltage source | Match |
| ParallelRLCSINCurrent | Parallel resistor, inductor and capacitor response to a current source | Match |
| ParallelRLDC | Parallel resistor and inductor response to a voltage source | Match |
| ParallelRLDCCurrent | Parallel resistor and inductor response to a current source | Match |
| ParallelRLPULSE | Parallel resistor and inductor response to a voltage source | Match |
| ParallelRLPULSECurrent | Parallel resistor and inductor response to a current source | Match |
| ParallelRLSIN | Parallel resistor and inductor response to a voltage source | Match |
| ParallelRLSINCentered | Parallel resistor and inductor response to a voltage source | Match |
| ParallelRLSINCurrent | Parallel resistor and inductor response to a current source | Match |
| ParallelRPULSE | Parallel resistor response to a voltage source | Match |
| ParallelRPulseCurrent | Parallel resistor response to a current source | Match |
| ParallelRSIN | Parallel resistor response to a voltage source | Match |
| ParallelRSINCurrent | Parallel resistor response to a current source | Match |
| SeriesCapDC | Adding capacitors in series with a voltage source | Match |
| SeriesCapDCCurrent | Adding capacitors in series with a current source | Match |
| SeriesCapPulse | Adding capacitors in series with a voltage source | Match |
| SeriesCapPULSECurrent | Adding capacitors in series with a current source | Match |
| SeriesCapSIN | Adding capacitors in series with a voltage source | Match |
| SeriesCapSINCurrent | Adding capacitors in series with a current source | Match |
| SeriesCurrentSourceAdditionDC | Intentionally failing adding current sources in series | BioGears: successfully fails - provides feedback on failure |
| SeriesCurrentSourceAdditionPULSE | Intentionally failing adding current sources in series | BioGears: successfully fails - provides feedback on failure |
| SeriesCurrentSourceAdditionSIN | Intentionally failing adding current sources in series | BioGears: successfully fails - provides feedback on failure |
| SeriesIndDC | Adding inductors in series with a voltage source | Match |
| SeriesIndDCCurrent | Adding inductors in series with a current source | Match |
| SeriesIndPULSE | Adding inductors in series with a voltage source | Match |
| SeriesIndPULSECurrent | Adding inductors in series with a current source | Match |
| SeriesIndSIN | Adding inductors in series with a voltage source | Match |
| SeriesIndSINCurrent | Adding inductors in series with a current source | Match |
| SeriesPressureSourceAdditionDC | Adding voltage sources in series | Match |
| SeriesPressureSourceAdditionPULSE | Adding voltage sources in series | Match |
| SeriesPressureSourceAdditionSIN | Adding voltage sources in series | Match |
| SeriesRCDC | Series resistor and capacitor response to a voltage source | Match |
| SeriesRCPULSE | Series resistor and capacitor response to a voltage source | Match |
| SeriesRCSIN | Series resistor and capacitor response to a voltage source | Match |
| SeriesRDC | Series resistors response to a voltage source | Match |
| SeriesRLCDC | Series resistor, inductor and capacitor response to a voltage source | Match |
| SeriesRLCDCCurrent | Series resistor, inductor and capacitor response to a current source | Match |
| SeriesRLCPULSE | Series resistor, inductor and capacitor response to a voltage source | Match |
| SeriesRLCPULSECurrent | Series resistor, inductor and capacitor response to a current source | Match |
| SeriesRLCSIN | Series resistor, inductor and capacitor response to a voltage source | Match |
| SeriesRLCSINCurrent | Series resistor, inductor and capacitor response to a current source | Match |
| SeriesRLDC | Series resistor and inductor response to a voltage source | Match |
| SeriesRLDCCurrent | Series resistor and inductor response to a current source | Match |
| SeriesRLPULSE | Series resistor and inductor response to a voltage source | Match |
| SeriesRLPULSECurrent | Series resistor and inductor response to a current source | Match |
| SeriesRLSIN | Series resistor and inductor response to a voltage source | Match |
| SeriesRLSINCurrent | Series resistor and inductor response to a current source | Match |
| SeriesRPULSE | Series resistors response to a voltage source | Match |
| SeriesRSIN | Series resistors response to a voltage source | Match |
| SimpleDiodeDC | Diode behavior in response to a voltage source | Match |
| SimpleDiodePULSE | Diode behavior in response to a voltage source | Match |
| SimpleDiodeSIN | Diode behavior in response to a voltage source | Match |
| SwitchRCDC | Circuit behavior when a switch is thrown with a voltage source | Match |
| SwitchRCDCCurrent | Circuit behavior when a switch is thrown with a current source | Match |
| SwitchRCPULSE | Circuit behavior when a switch is thrown with a voltage source | Match |
| SwitchRCPULSECurrent | Circuit behavior when a switch is thrown with a current source | Match |
| SwitchRCSIN | Circuit behavior when a switch is thrown with a voltage source | Match |
| SwitchRCSINCurrent | Circuit behavior when a switch is thrown with a current source | Match |
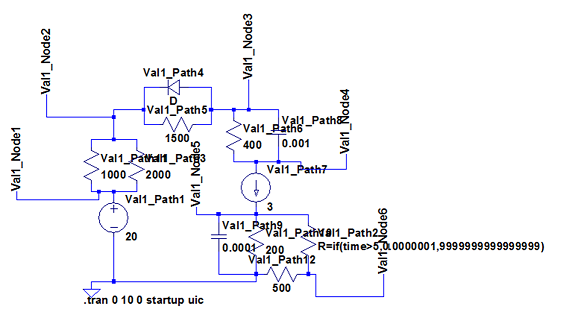
The most interesting and complex circuits use a combination of all elements. Two of these comprehensive circuits are shown below because of their complexity. The first uses sine wave sources and the second uses pulse sources. There is a strong correlation between the given LTspice outputs and the calculated BioGears outputs. Note that the sign convention of the current across voltage sources is reversed for LTspice, because it does not maintain the source-to-target positive current standard as is done with BioGears.
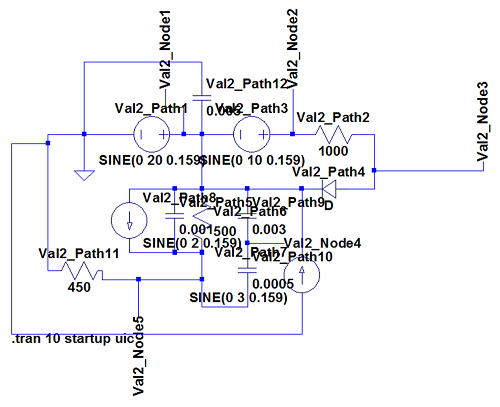
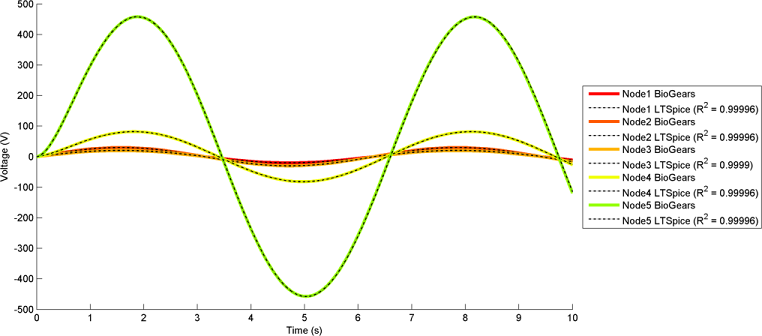
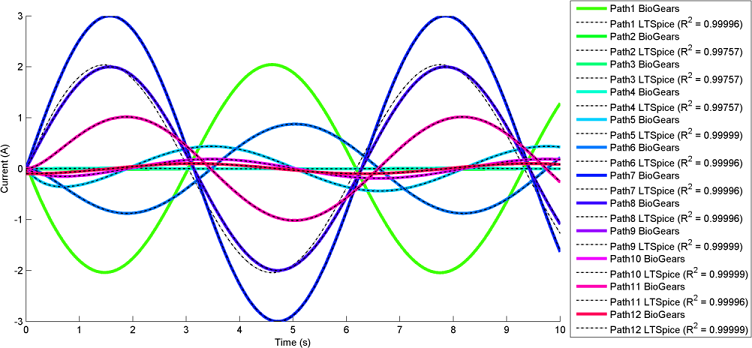
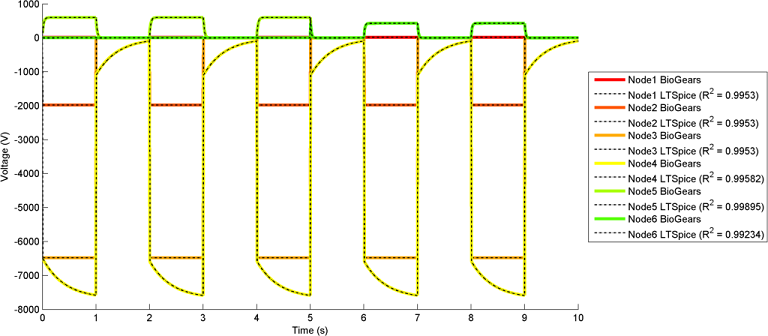
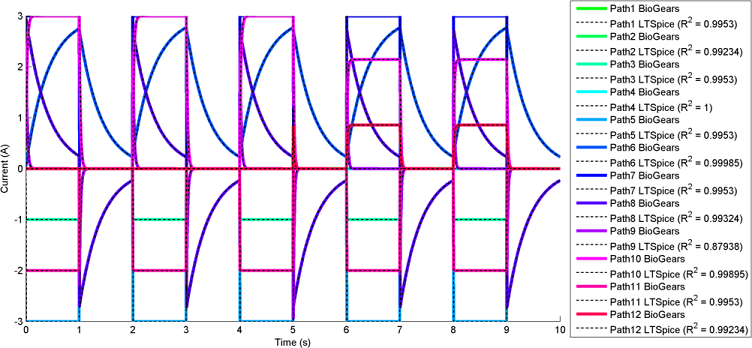
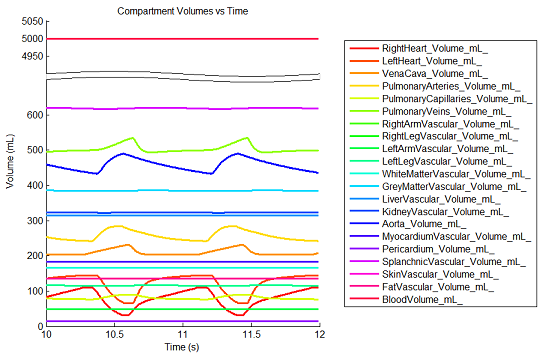
BioGears has been shown to successfully conserve mass, energy, and momentum within all defined closed-loop systems. The successful conservation of mass provided by the solver is shown in Figure 10. The volume (quantity/charge) within cardiovascular circuit nodes through approximately 2.5 full heart beat cycles. The total volume of all compartments remains at a constant value of 5L throughout the entire process.
All basic Circuit Solver functionality is further validated and verified with specific unit tests that target individual methods. The following functionality has been successfully validated by individual tests:
- Incorrect usage errors
- Combined circuits
- Dynamically-changing elements
- Dynamically-changing circuits
- Added and removed elements
- Fluid, thermal, and electrical model types
- Non-zero reference node voltage
- Polarized elements
- Dynamically modifying capacitor charge
Conclusion
The BioGears generalized circuit analysis techniques have successfully accomplished all of the goals for that area of the code. The solver implements the CDM effectively and generically in a way that allows for infinitely complex lumped parameter models that can also be combined. We made it simple to modify existing functionality and add new models. This has saved, and will continue to save, significant amounts of development time. The generic and modular design also allows for much easier bug finding and fixing, system parameter tuning, and system validation. The burden for the modeler is on creating a good model and not worrying about implementing sound physics math.
The Circuit Solver has also been designed such that it can be extracted from BioGears and used for any software that needs to perform transient circuit analysis.
Future Work
Coming Soon
The list below includes some of the planned functionality additions.
- Functionality to create port connections between circuits for improved modularity between systems/subsystems
Recommended Improvements
Other functionality that may be beneficial includes:
- A sensitivity analysis and error term determination
- An analysis of frequency constraints
- More circuit elements (i.e., amplifiers, transformers, transistors, non-ideal elements, zener diodes, etc.)
Appendices
Data Model Implementation
SECircuit
SECircuitCalculator
SECircuitNode
Acronyms
CDM - Common Data Model
DC - Direct Current
GUI - Graphical User Interface
KCL - Kirkoff's Current Law
MNA - Modified Nodal Analysis
SPICE - Simulation Program with Integrated Circuit Emphasis





