The Common Data Model (CDM) is an Object Oriented Design (OOD) of class structures that provides a unified set of data encapsulation objects that will promote fast development, compatible data sets, and well-defined interfaces for all types of physiology model development. The CDM provides a well-defined data interchange format that disparate models can use for standardizing inputs and outputs, making it easy for physiology models and engines to be extended and operate jointly. The CDM is I/O indifferent; it provides the ability to hold all data associated regardless of the data being an input or an output to a specific model. These structures provide discrete properties for specific definitions and the flexibility to be easily expandable when incorporating new models. The CDM classes are small, lightweight (not much code in them), property-bag objects intended to only contain data for simple I/O and message passing.
You can access the CDM data dictionary via the modules link provided in the above header. You can also view Common Data Model Tables associated with some of key objects associated with the Physiology Engine Interface . This dictionary documents the conceptual structure, organization, relationships, and property definitions. A set of base types, known as Properties, is defined to hold the actual discrete values of various objects. The CDM then encapsulates and organizes these properties into various physiology-related data objects, such as Systems, Anatomy, Equipment, Patients, Actions, etc.
The CDM data dictionary is implemented in an eXtensible Markup Language (XML) Schema Description (XSD). An XML schema is an open standard that describes the structure of an XML document and allows for defining data that programs can process in a generic manner. This representation enforces the meaning and structure and relationships as described in the data dictionary and provides an industry standard basis for implementation and data exchange. This XML schema can then be used to generate binding classes that can serialize XML for use in programming languages such as C++, Java, and C#.
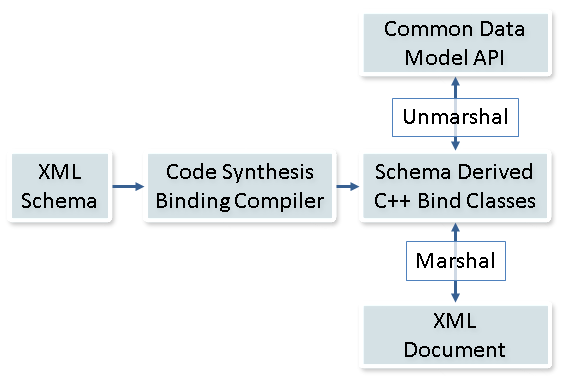
We then created a set of API classes built to reflect the organization of the data dictionary, encapsulate the binding classes, and provide various data validation, manipulation, and translation algorithms. These classes are prefixed with SE, which stands for Synthetic Environment. These classes provide an encompassing interface for holding and translating data in a simple-to-use, yet flexible and expandable, interface. This library will provide application developers with low-level infrastructure (serialization, translation, data reuse, etc.) already complete so they can concentrate on creating end-user content-driven applications.
Properties
An SEProperty is an object that represents a basic data type in the CDM that stores the actual data.
There are several Property data types:
- SEScalar - A combination of a double and a unit. The CDM does not require a property in a specific unit of measure (for example, the height of a patient does not have to be in meters). Rather, the CDM requires each data type to retain a unit of measure of the Property's quantity and the class will translate the value to a requested unit. One example is a Distance Property that holds data in any distance-associated unit: meters, feet, inches, etc.
- Strings - We use the inherent language type for string
- Enumeration - Where applicable, instead of strings, we use enumerations for code clarity and speed.
- SEFunction - A combination of an abscissa and an ordinate paired together.
Modeling Objects
Modeling objects are hierarchically organized data containers based on the needs of physiology modeling and simulation. These objects contain data by holding specifically named Property objects. They can also hold other modeling types to achieve the proper hierarchy of data organization. Since these objects do not contain the actual data values (they contain Properties) and optionally other modeling objects, their interface is based on a get/has style. When you call a get method (e.g., getVolume or getRightHeart) on an object, the requested object or property is returned (created on demand if it did not previously exist) (e.g., SEScalarVolume or SEHeart). The complementing has method (e.g., hasVolume or hasRightHeart) is a test to show if the object already exists or not.
CDM Algorithms
A few generic algorithms are available in the CDM. The following sections describe some of these algorithms.
Unit Conversion
The Common Data Model has a unit conversion library built into the SEScalar class that will convert double data to whatever unit you specify. There is an SEScalar class for various quantity types along with commonly used unit definitions in those classes (such as SEScalarLength has units : m, cm, in, etc.) You may use strings or a CCompoundUnit to define your unit if you do not find a unit predefined, but strings have a runtime cost to them. Units are defined in the standard abbreviation, as described at: http://www.bipm.org/en/si/. Along with SI units, English units and medical units are supported. You can check to see if a unit is valid by calling the IsValidUnit method on a particular quantity scalar class.
Logging
The Data Model has a logging capability that each SE class uses to log any info to a log file. This logger has different levels of logging an event:
- Debug - Extremely low level information, intended for the developer to assess any questions in functionality
- Info - A general level of information where the engine wants to inform a user of something
- Warning - Something was not inputted properly or an assumption was made in the underlying methodology
- Error - A recoverable error occurred and the engine is still able to proceed, but results should be inspected for correctness
- Fatal - A unrecoverable error occurred and the engine will stop calculating
The logger allows a user to programmatically add an object to the logging class that is called when a log event occurs. This allows a calling program to be notified so that they may react to these logs. (e.g., error handling)
Data Track
A Data Track class is used for debugging purposes. Usually in debugging, one likes to print values of variables to the screen. This class allows a developer to write those values to a comma-delimited, time-ordered file. Developers can add any number of variables to track (only limited to machine memory) and they will be added to the output file for each time step.
Circuit Solver
The Circuit objects have algorithms for solving any closed-loop circuits as described in the Circuit Methodology.
Substance Transport
Substances are contained on compartments and are transported around the body based on volumes, pressures, and flows, which is handled via a generic Substance Transport Methodology.
Compartments
A compartment can represent various fidelities of data; it can be:
- An anatomical space, such as the body's skin
- An organ, such as the liver
- An organ substructure, such as the Left Heart
- A component of a piece of equipment, such as an anesthesia machine ventilator
Compartments are intended to be a generic data exchange interface between systems for the fluid dynamics data of the body, such as volumes, pressures, and flows. In addition to the fluid dynamics, compartments also contain a substance quatity for each substance in the compartment. For example, a vascular compartment includes the masses and concentrations of all substances in that compartment, whereas a pulmonary compartment will contain the volumes and volume fractions of all substances in that compartment. This generic implementation allows any system to access and modify the parameters for the fluid dynamics calculations reguardless of the model type.
We have a implemented a compartment interface to integrate directly with the circuit CDM classes to easily access the circuit-based fluid dynamics data. These circuit compartments are assigned nodes and paths and combine that data to provide an accurate fluid dynamics data set for a specified compartment.
The circuit compartment fluid dynamics data is combined in the following manner:
- Volume : Sum of the volume of all nodes in the compartment.
- Pressure : Pulled from a single node of a compartment that was explicitly identified during setup.
- Flow : The sum of inflows; if the inflow equals zero, it is the sum of the outflows.
- Vascular Substance Quantites
- Mass - The sum of all the masses for each node in the compartment for a particular substance
- Concentration - The compartment's substance mass divided by the compartment volume
- Pulmonary Substance Quantites
- Volume - The sum of all the volumes for each node in the compartment for a particular substance
- Volume Fraction - The compartment's substance volume mass divided by the compartment volume





